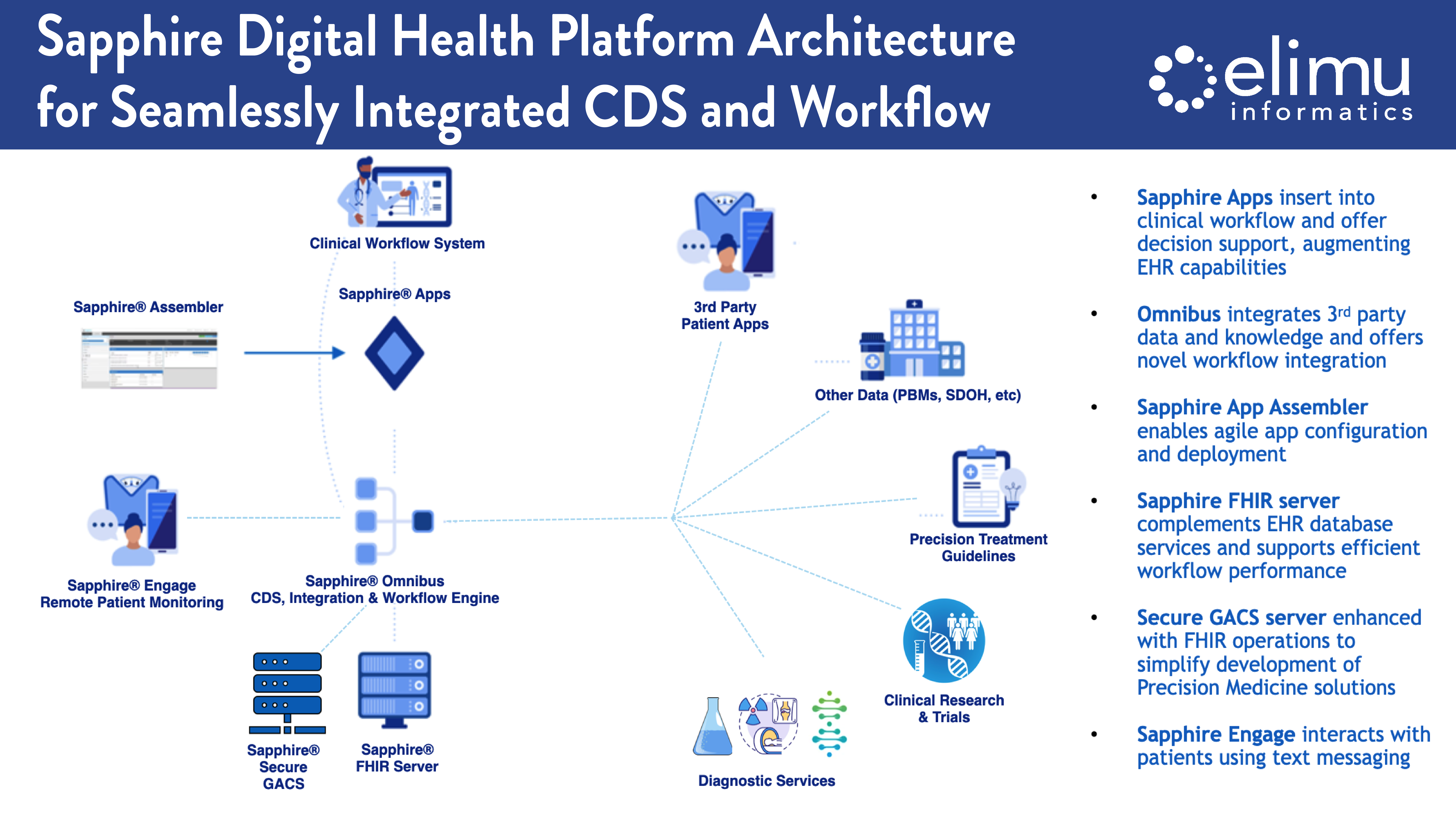
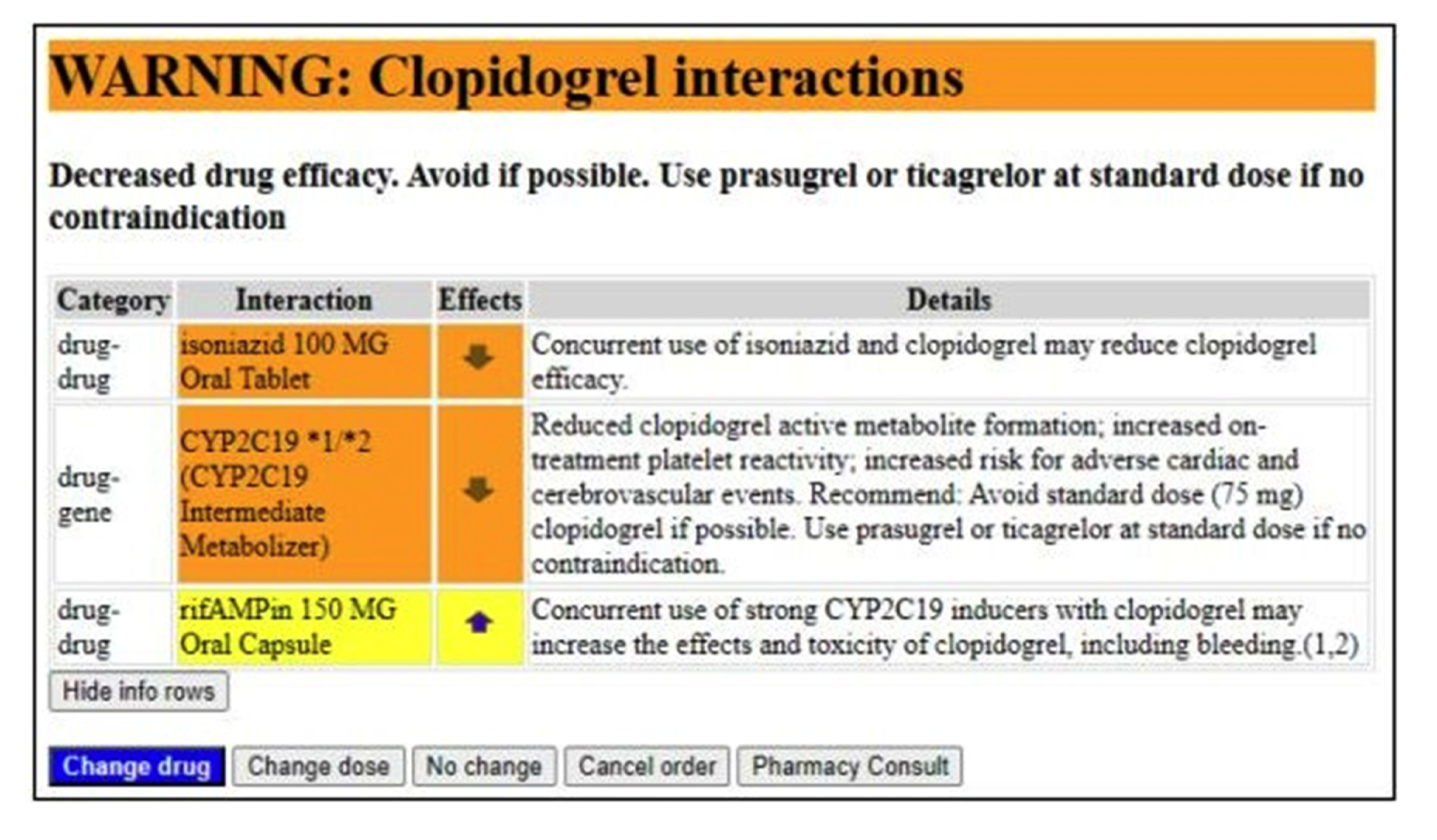
Research and Development: Precision Medicine
Secure Genome Archiving and Communication System (Secure GACS) Enables Secure, Robust Genomic Data Accessibility for Clinical Decision Support
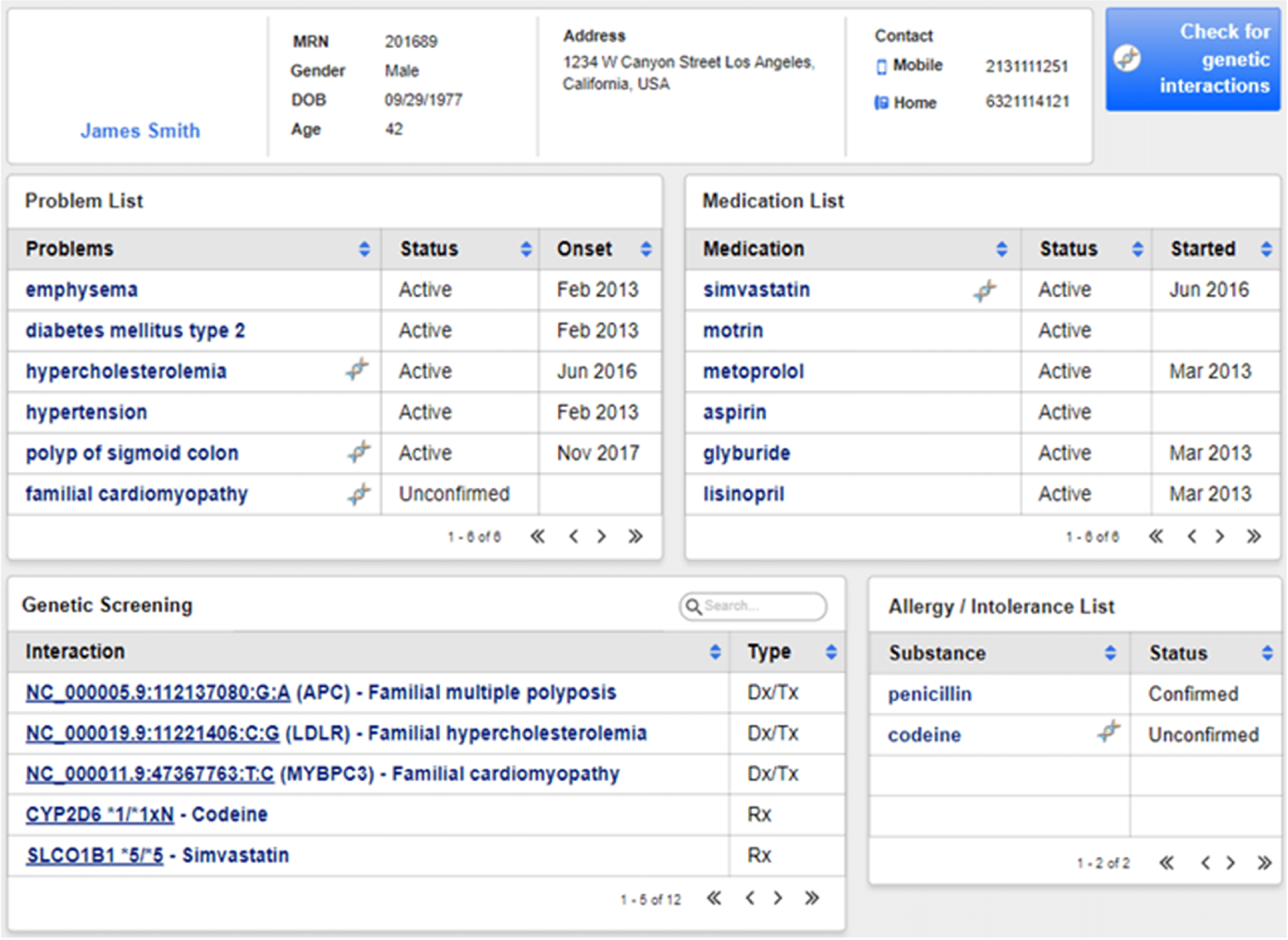
Genome data are large, comprising billions of base-pairs on thousands of genes and intergenic regions. Next-generation sequencing can identify up to millions of variants, whose clinical significance can change over time as our knowledge evolves. Sequencing can produce gigabytes of data for a single individual. It is impractical to securely store and analyze such large data in contemporary electronic health record (EHR) systems, which clinicians use when delivering care to patients. The challenges for storage can be more acute for smaller healthcare facilities that may not have large, secure data repositories. This means that genomic data must be stored outside the EHR system and retrieved for clinical decision support (CDS) purposes. Elimu Informatics researchers collaborated in the development of a prototype genomic archiving and communications system to securely store genome data and provide clinical decision support. This system operates on a client-server model. The client encrypts the data, and the server stores data and performs the computations necessary for CDS. Computations are directly performed on encrypted data, and the client decrypts results. The server cannot decrypt inputs or outputs, which provides strong guarantees of security. The team validated our system with three genomics-based CDS applications. The results demonstrated that it is possible to resolve a long-standing dilemma in genomic data privacy and accessibility, by using a principled cryptographical framework and a mathematical representation of genome data and CDS questions.1
Encapsulating the Complexity of Molecular Data with Standard FHIR Genomics Operations (APIs) so that Precision Medicine Solution Developers Can Focus on Building Clinical Decision Support Applications
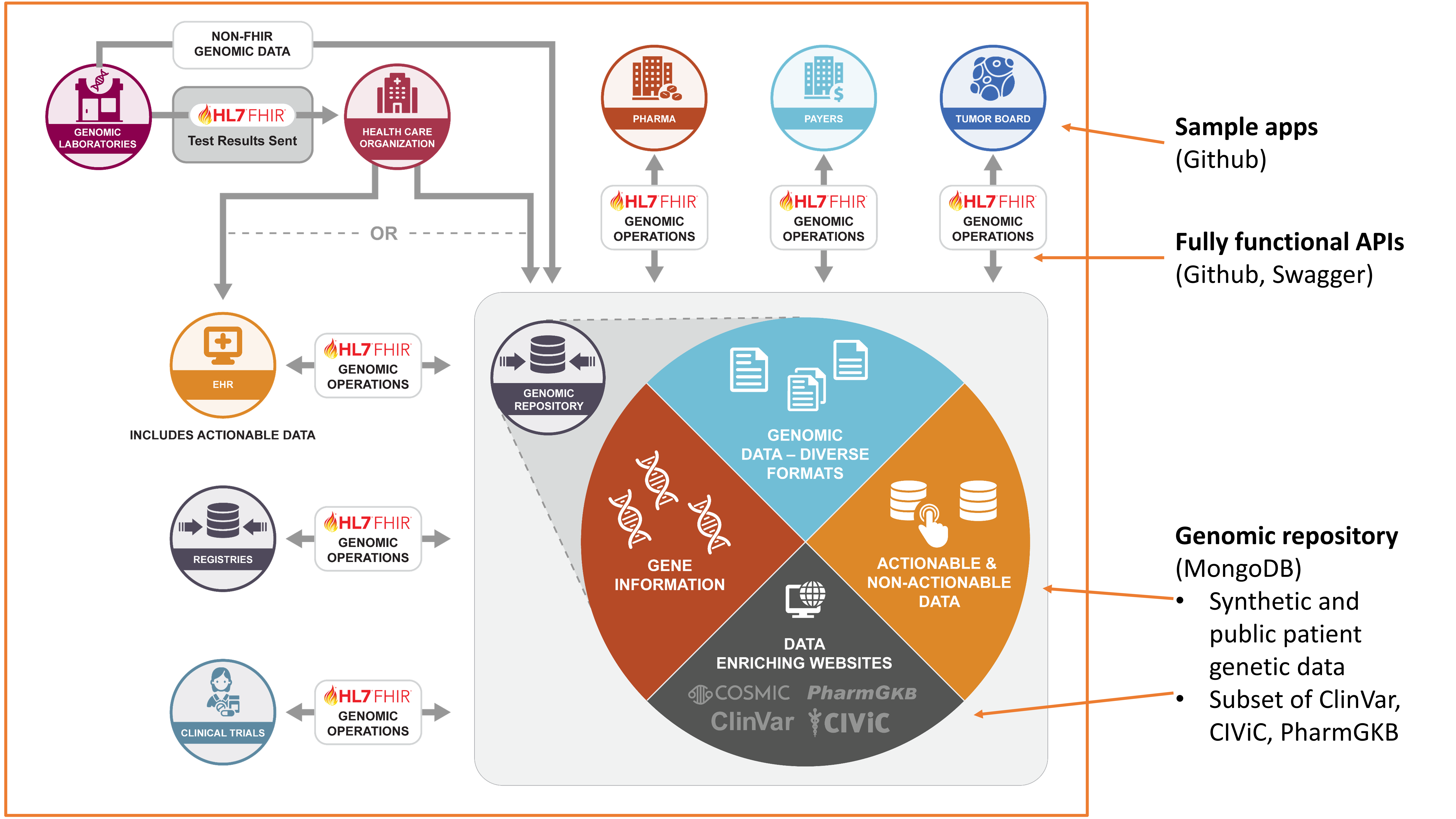
Click image to enlarge.
Figure: (Image courtesy of HL7 FHIR Accelerator CodeX™ from GenomeX site, (github | swagger).
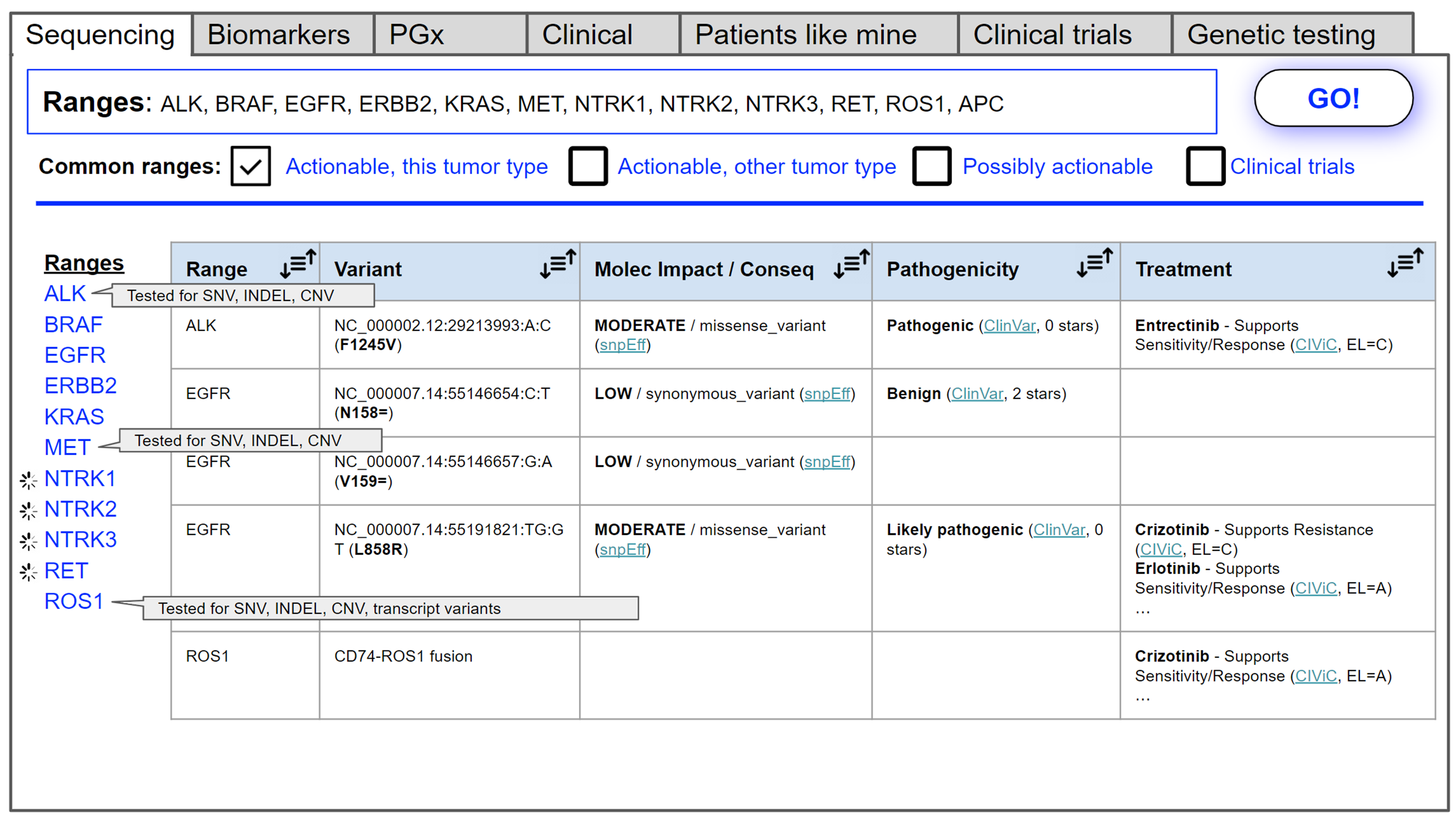
Next-generation sequencing can identify thousands to millions of variants, whose clinical significance can change over time as our knowledge evolves. To manage such a large volume of dynamic and complex data, new models of genomics-EHR integration are needed. Qualitative observations to date suggest that freeing application developers from the need to understand the nuances of genomic data, and instead base applications on standardized APIs can not only accelerate integration but also dramatically expand the applications of Omic data in driving precision care at scale for all. The Elimu Informatics research team and collaborators have proposed a new paradigm in genomics-electronic health record (EHR) integration, using a standardized suite of FHIR Genomics Operations that encapsulates the complexity of molecular data so that precision medicine solution developers can focus on building applications. Fifteen FHIR Genomics Operations have been developed, designed to support a wide range of clinical scenarios, such as variant discovery; clinical trial matching; hereditary condition and pharmacogenomic screening; and variant reanalysis. Operations are currently being matured through the HL7 balloting process, connectathons, pilots, and the HL7 FHIR Accelerator program.2
Participation in GenomeX, a Standards-development Collaboration Effort Under the HL7 FHIR Accelerator, CodeX, That Can Help Innovate Clinical Trials Matching and Molecular Tumor Board
In May of 2022, HL7 launched GenomeX to facilitate interchange of machine-readable genomics data following the FHIR specification. GenomeX is housed within an HL7 FHIR accelerator called CodeX, a community of individuals and organizations with an interest in interoperability of all kinds of health data. Elimu Informaticians are key contributors to this group that includes representatives from molecular laboratories, care providers, payers, technology vendors, and patients.
(Genomic Data Standards Work Inches Forward With New HL7 GenomeX Initiative, CodeX, HL7 International Launch GenomeX Community to Enhance Access to Genomic Data for Improved Patient Care)
Streamlining Genomics Data Integration with the Electronic Health Record
While VCF formatted files are the lingua franca of next-generation sequencing, most EHRs do not provide native VCF support. As a result, labs often must send non-structured PDF reports to the EHR. On the other hand, while FHIR adoption is growing, most EHRs support HL7 interoperability standards, particularly those based on the HL7 Version 2 (HL7v2) standard. The Elimu Informatics team collaborated first on a project that demonstrated how an open source vcf2fhir utility can enable conversion of variants from VCF format into HL7 FHIR format via two case studies. The first use case, 'SMART Cancer Navigator', is a web application that offers clinical decision support by linking patient EHR information to cancerous gene variants. The second, 'Precision Genomics Integration Platform', intersects a patient's FHIR-formatted clinical and genomic data with knowledge bases in order to provide on-demand delivery of contextually relevant genomic findings and recommendations to the EHR. A later project focused on extension of the utility to output HL7v2 LRI data, a standard interoperability data standard for integrating lab results into an electronic health record. This extension contains both variants and variant annotations (e.g., predicted phenotypes and therapeutic implications). Using this HL7v2 converter, the team implemented an automated pipeline for moving structured genomic data from the clinical laboratory to the EHR. The team also developed an open source hl7v2GenomicsExtractor that converts genomic interpretation report files into a series of HL7v2 observations conformant to HL7v2 LRI.3
Enabling Continuous Genomic Variant Reanalysis to Ensure Most Current Interpretation for Clinical Decision Support
Variant annotation is crucial in next-generation sequencing, allowing labs to sift through numerous variants and pinpoint the most relevant ones, offering clinicians vital context for decision-making. However, as genomics knowledge evolves rapidly, reported annotations can swiftly become outdated. Variants of unknown clinical significance can quickly become understood to have significant import for the patient and it is essential to have mechanisms that enable surveillance and notification of clinicians and patients alike of these evolving interpretations.
Under the ONC Sync for Genes program, the Elimu Informatics team and collaborators sought to standardize the sharing of dynamically and continuously annotated variants (e.g., variants annotated on demand, based on current knowledge). The computable biomedical knowledge artifacts that were developed enable a CDS application to surface up-to-date annotations to clinicians. Using standards for HL7 FHIR Genomics and Global Alliance for Genomics and Health (GA4GH) Variant Annotation (VA), the team developed a CDS pipeline that dynamically annotates patient's variants, providing FHIR-encoded variants and annotations (diagnostic and therapeutic implications, molecular consequences, population allele frequencies) via FHIR Genomics Operations. ClinVar, CIViC, and PharmGKB were used as knowledge sources, encoded as per the GA4GH VA specification.4
- Karimi S, Jiang X, Dolin RH, Kim M, Boxwala A. A secure system for genomics clinical decision support. J Biomed Inform. 2020 Dec;112:103602.
- Dolin RH, Heale BSE, Alterovitz G, Gupta R, Aronson J, Boxwala A, Gothi SR, Haines D, Hermann A, Hongsermeier T, Husami A, Jones J, Naeymi-Rad F, Rapchak B, Ravishankar C, Shalaby J, Terry M, Xie N, Zhang P, Chamala S. Introducing HL7 FHIR Genomics Operations: a developer-friendly approach to genomics-EHR integration. J Am Med Inform Assoc. 2023 Feb 16;30(3):485-493.
- Dolin RH, Gothi SR, Boxwala A, Heale BSE, Husami A, Jones J, Khangar H, Londhe S, Naeymi-Rad F, Rao S, Rapchak B, Shalaby J, Suraj V, Xie N, Chamala S, Alterovitz G. vcf2fhir: a utility to convert VCF files into HL7 FHIR format for genomics-EHR integration. BMC Bioinformatics. 2021 Mar 2;22(1):104. and Dolin RH, Gupta R, Newsom K, Heale BSE, Gothi S, Starostik P, Chamala S. Automated HL7v2 LRI informatics framework for streamlining genomics-EHR data integration. J Pathol Inform. 2023 Aug 15;14:100330.
- Dolin R, Heale BSE, Gupta R, Alvarez C, Aronson J, Boxwala A, Gothi SR, Husami A, Shalaby J, Babb L, Wagner A, Chamala S. Sync for Genes Phase 5: Computable artifacts for sharing dynamically annotated FHIR-formatted genomic variants. Learn Health Syst. 2023 Aug 30;7(4):e10385.
- Dolin RH, Boxwala A, Shalaby J. A Pharmacogenomics Clinical Decision Support Service Based on FHIR and CDS Hooks. Methods Inf Med. 2018 Dec;57(S 02):e115-e123.