Jan 29, 2024 | Dr. James Shalaby
AI is everywhere now and continues to permeate into every aspect of our lives.
Value sets are collections of encoded concepts that help us achieve semantic interpretation, normalization and summarization of patient data. As an expert terminologist with decades of experience curating value sets for a myriad of clients, I decided to explore this question a little further with just the tools available online today. In summary I am impressed with the potential of today’s AI engines to reduce the labor AND I did find some limitations that will likely be addressed in the near future as well as other limitations that I’m just not sure if AI will be able to address any time soon.
I focused on exploring how well two different engines perform with respect to creating value sets in the medications and laboratory domains. I chose these domains because (a) they are well defined (man-made “synthetic” domains) and (b) they are very easy for me (a pharmacist) to validate.
Value sets define useful, reusable lists of concepts such as test results, conditions, medication, findings and procedures that may be needed for specific purposes. They allow systems to define an entity as a kind of broader concept such as which code or named procedure indicates a total hysterectomy. They can also be utilized to define a set of relationships to an entity such as which code or named conditions, medications, test results, and findings might be an indication of immunosuppression. Value sets are often, in today’s world, a foundational substrate for the development of clinical decision support algorithms embedded in clinical workflow systems such as electronic health records.
For example, a clinical decision support algorithm would use value sets to reason over a patient chart and determine if a patient has a history of myocardial infarction and if this patient is prescribed a beta-blocker. They are also a substrate for analytic and research queries looking to define cohorts of patients by similarly specified criteria. Due to the variety and complexity of standard terminologies, the manual curation of value sets can be extremely labor intensive. Today’s publicly available value sets are not always robustly maintained hence my professional services team is continuously engaged in value set curation projects.
In this exploration of the potential for AI to reduce the dependency on manual curation, I defined success criteria as value sets that are conceptually accurate but can have gaps in content. For example, if a value set for systemic beta blockers contained all the right RxNorm ingredients and most/many of the formulations but had some minor gaps in certain strengths or combinations I considered that a success. I also defined failure criteria as a value set that contained incorrect member concepts or major gaps of omission. Again, for the same example if it also contained beta agonists, alpha adrenergic blockers, calcium channel blockers , these would all constitute failures unless they were in combination with a beta blocker.
The reason behind my less than perfect acceptance of value sets success criteria is that it’s much easier for a terminologist to expand on a candidate value set to fill in gaps than it is to find and remove erroneous inclusions.
Findings:
Generally the value sets generated are more useful to a terminologist as an aid in creating final versions that incorporate the exhaustive scope of codes needed to support most decision support. However, they would not be appropriate for direct use without further editing and curation.
1. Question to Bard and ChatGPT (simplest example):
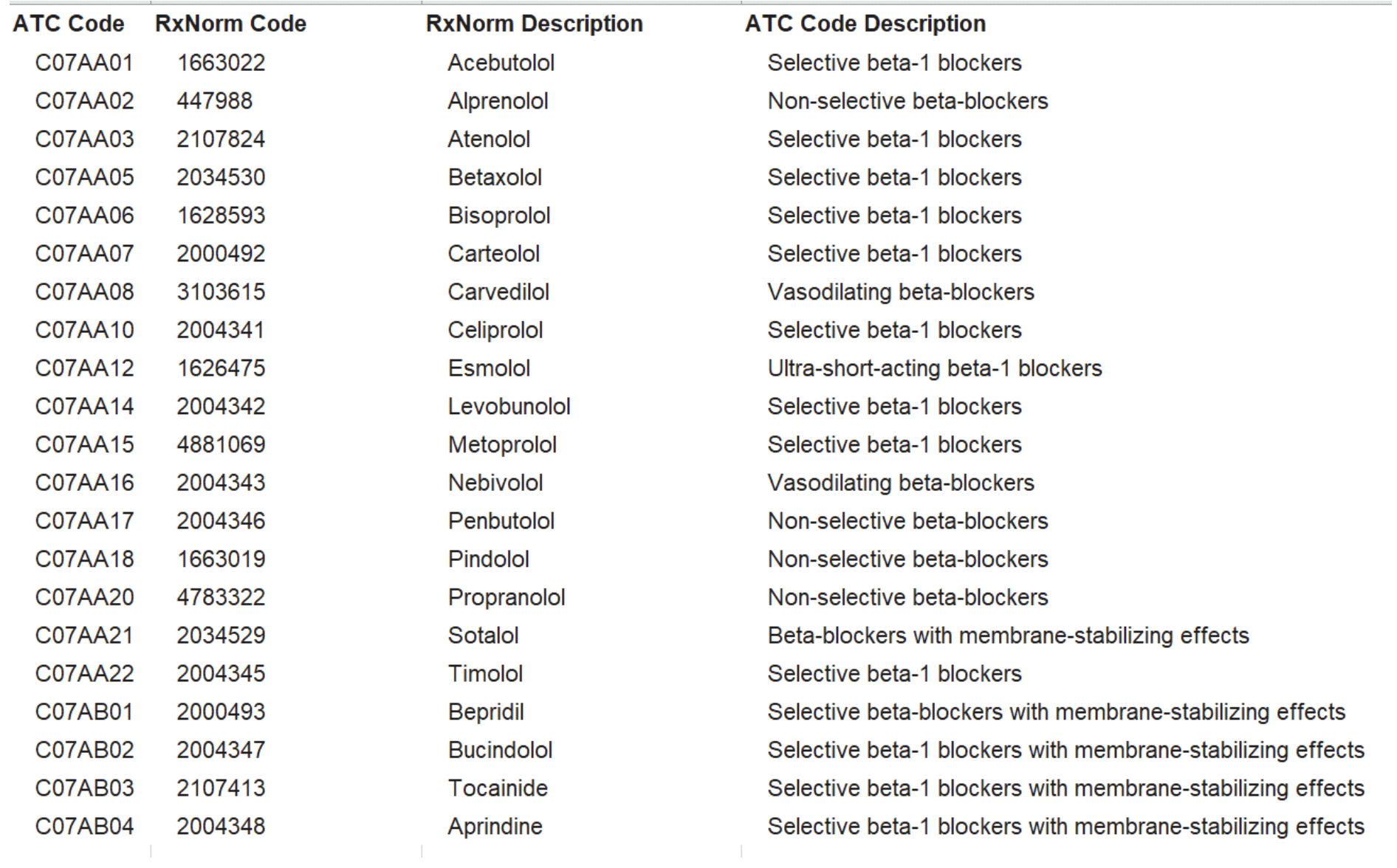
“Create an rxnorm value set of all systemic beta blockers using Anatomical Therapeutic Chemical (ATC) classifications and show the ATC codes used to create the value set as well as the rxnorm codes”
“Include descriptions for the ATC classes and the rxnorm codes”
Results:
It returns a nice tabular response that has the rxnorm codes and descriptions as well as the corresponding ATC classes. However, there are gaps at 2 levels:
Conceptual gaps:
It missed some rxnorm beta blockers conceptually completely. It was minor and traceable partially perhaps to gaps in ATC data (ATC classifies rxnorm ingredients.
Formulation gaps (rxnorm term type gaps):
Both methods could only return ingredient level concepts. It could not return the more specific rxnorm codes for different strengths and dose forms (formulation level) and definitely could not return brand names (which was really not an issue from my perspective).
2. Question:
“Create an intensional rule for the above using ATC”
Results:
Bard nicely defines in words each rule to define this value set which is a very helpful feature. However, the steps are only partially correct and will not produce the results that a terminologist would expect. The intensional rules from Bard are shown below. First mistake: The ATC to RXNORM mappings actually do exist in RXNORM and have for many years. Second mistake is that it made the assumption that the term types of interest are GPCK (generic pack level such as birth control pills) and SCD (semantic clinical drug, a non-branded formulation description such as simvastatin 10mg oral tablet) but it really should have included SCDG (semantic clinical drug group, a non strength specific representation of a drug such as simvastatin oral product) and SCDF (semantic clinical drug form, a dose form specific abstraction without strength such as simvastatin oral tablet) as well for the assumption it made that this is a medications value set and not a medication allergies value set (which would have just been IN (ingredient) and MIN (multi-ingredient) term types (ingredients).
ChatGPT 3.5 produced a less useful but partially correct representation. It’s possible that ChatGPT 4.0 would have performed better but the results were not as useful as Bard. An example of the results are shown below. Interestingly, ChatGPT attempted to actually “write code” for the intensional rule (albeit incorrect and not really code but markdown).
Conclusions:
- The above exercise illustrates the cautions that we should exercise not to fully depend on Large Language Model (LLM) methods to infer data about patients. If we were looking to implement a clinical decision support algorithm ensuring that candidate patients are appropriately receiving beta blockers, an AI derived algorithm would potentially erroneously suggest a patient is not receiving one which would be annoying to the clinician recipient of the suggestion. Or, if a researcher or quality reporting analyst is utilizing an LLM query to identify all patients who are candidates for beta blockers, but not receiving them, such a query would harvest a cohort of patients who are actually on them.
- AI has a lot of potential as a tool for helping terminologists more efficiently produce and maintain value sets. I can imagine huge benefits on initial creation as well as facilitation of maintenance once the value sets are created.
- Currently I would not trust AI to understand a well formed prompt (question to Bard or ChatGPT) and produce quality that is close to what even an average terminologist would produce. It simply does not understand how to incorporate context of use of the value set into its design at this time.
- AI does have the potential of helping with value set searches , doing value set comparisons, and even providing candidate feedback loops to reference terminologies used in intentionally defined value sets. This last point is interesting since value sets that for example incorporate concepts that define diabetes mellitus through multiple hierarchies in SNOMED CT may potentially identify inconsistencies in the terminology hierarchies.
- Generally, AI at its current level of performance should not be used to automate generation of medication value sets for direct use in CDS or analytics. I’m certain that over time it’ll improve significantly but for now it does require human validation.
Elimu Informatics provides advisory services for artificial intelligence solution evaluation and deployment as well as services for standard terminology mapping, value set curation, and clinical decision support rule authoring to our customers in all healthcare verticals. Please feel free to contact me at info@elimu.io if you would like to explore these possibilities in more detail or might have a need for such content engineering services.
Bard results:
Intensional Rules for “Beta Blockers (RxNorm)” Value Set
This section defines the intensional rules for the “Beta Blockers (RxNorm)” value set, allowing it to dynamically update as new medications are added or removed.
Rule 1:
Concept type: GPCK (Generic Product) OR SCD (Substance)
Reasoning: This rule ensures the value set only includes medication concepts, not individual ingredients or product forms.
Rule 2:
Concept name: Contains one of the following keywords:
Acebutolol
Atenolol
Betaxolol
Bisoprolol
Carvedilol
Esmolol
Labetalol
Metoprolol
Nadolol
Penbutolol
Pindolol
Propranolol
Sotalol
Timolol
OR
Concept name: Contains the term “beta blocker” (case-insensitive)
Reasoning: This rule identifies concepts representing beta-blocker medications by searching for specific keywords or the general term.
Rule 3:
Concept marketing status: Currently marketed in the United States
Reasoning: This rule ensures the value set includes only medications actively used in the US healthcare system.
Rule 4:
Exclude concepts with the following RxNorm Type TTYs:
IN (Ingredient)
MPN (Multi-part Product)
UNII (Unique Ingredient Identifier)
Reasoning: This rule excludes individual ingredients, multi-part products, and unique ingredient identifiers to focus solely on medications.
Rule 5:
Update frequency: Monthly
Reasoning: This rule ensures the value set is reviewed and updated monthly to reflect any changes in the RxNorm database, including new medications or discontinuations.
These intensional rules allow the “Beta Blockers (RxNorm)” value set to dynamically adapt to changes in the medication landscape while maintaining its accuracy and relevance.
profile picture
use ATC for that intensional rule
View other drafts
Unfortunately, I cannot directly use ATC (Anatomical Therapeutic Chemical Classification System) for the intensional rule of the “Beta Blockers (RxNorm)” value set. While ATC is a valuable tool for classifying medications, it does not directly map to RxNorm codes.
However, we can still use ATC to enhance the rule by excluding medications with ATC codes outside the beta-blocker category. This would be an additional rule to the existing ones:
Rule 6:
Exclude concepts with the following ATC codes:
C07AA (Beta-blocking agents, plain)
C07AB (Beta-blocking agents, selective)
C07AG (Beta-blocking agents, vasodilating)
C07BB (Angiotensin II receptor antagonists, plain)
Reasoning: These ATC codes represent medications that are not classified as beta-blockers. Excluding them ensures the value set remains focused on its intended target.
Combining the existing rules with this additional ATC-based rule will improve the specificity and accuracy of the “Beta Blockers (RxNorm)” value set.
ChatGPT 3.5 Results:
Value Set: Beta Blockers
Criteria:
– Drug class is “Beta Blockers”
– RxNorm code is one of the following: [List of RxNorm codes for beta blockers]
– ATC code starts with “C07” (indicating the cardiovascular system, beta-blocking agents)
Individual Drugs:
– Atenolol
– Drug class: “Beta Blockers”
– RxNorm code: [RxNorm code for Atenolol]
– ATC code: C07AB03
– Metoprolol
– Drug class: “Beta Blockers”
– RxNorm code: [RxNorm code for Metoprolol]
– ATC code: C07AB02
– Propranolol
– Drug class: “Beta Blockers”
– RxNorm code: [RxNorm code for Propranolol]
– ATC code: C07AA05
– Carvedilol
– Drug class: “Beta Blockers”
– RxNorm code: [RxNorm code for Carvedilol]
– ATC code: C07AG02