Mar 19, 2019 | Elimu Informatics
Over the past few years, Elimu has experimented with FHIR Genomics and we have actively participated in standards development activities. The January 2019 balloted FHIR Genomics guide is a major milestone. Once ballot comments have been resolved and applied (by summer 2019), we will have a remarkably useful guide for semantic interoperability of genomic findings and interpretations. The balloted version of FHIR Genomics can be successfully implemented against a number of real world use cases, to meet a number of genomics-EHR integration challenges such as pharmacogenomics, variant reanalysis, and patient-facing apps.
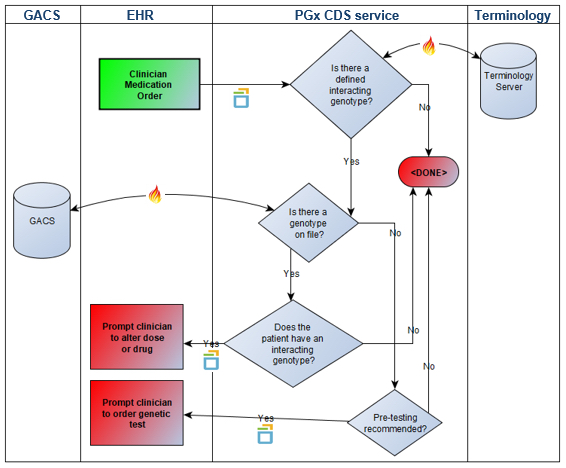
At the January 2019 HL7 FHIR Connectathon, we tested FHIR Genomics in a pharmacogenomics clinical decision support (PGx CDS) scenario. The scenario involved pushing PGx recommendations into Cerner EHR via our CDS-Hooks platform during the prescribing workflow. Genetic findings were obtained from a FHIR-enabled Genomic Archiving and Communication System (GACS) supplied by our friends at Intermountain Healthcare. The data flow is depicted in the following figure.
As part of this project, we created a VCF to FHIR converter, that takes variants identified via next-generation sequencing and stored in a Variant Call Format (VCF) file and converts them into a FHIR Diagnostic Report conforming to the January 2019 balloted FHIR Genomics guide. VCF to FHIR conversion facilitates incorporation of raw genomics data into EHR and CDS workflows, thus serving as a foundational component of Genomics-EHR integration strategies. As more and more consumers participate in gene sequencing ordered by their physicians or through Direct-to-Consumer testing centers, that data will be stored in a VCF format within large GACS repositories. Making that data available in FHIR format, which is gaining adoption across EHRs, will help make a person’s full genomic data accessible for precision health care.
We tested the converter against 17 anonymized VCF files obtained from the 1000 Genomes project. From each VCF file, we extracted gene-specific variants using bcftools, creating 3 gene-specific (TPMT, CYP2C19, CYP2D6) VCF files for each of 17 patients, for a total of 51 VCF files. Each of these files was then translated, via the VCF to FHIR converter, into a corresponding FHIR Genomics report. We’d like to make this converter, along with the generated FHIR instances, available to the community to help further Genomics-EHR integration exploration. A full description of the software, along with source code, source VCF files, and corresponding FHIR Genomics reports (in XML and JSON) can be found in our downloads section here.
While the current FHIR Genomics standard accommodates several key scenarios in precision medicine, additional development is required to tackle more complex workflows. We anticipate continually enhancing the code and resources, and we welcome the collaboration of other groups through that venue.