Mar 19, 2024 | Elimu Informatics
Our Vision for Precision Medicine
The notion of ‘precision medicine‘ or tailoring diagnostics and therapeutics based on individual patient characteristics isn’t new. What is new is the growing availability of patient-specific standardized data ranging from core EHR data (e.g. conditions, medications, allergies, labs) to molecular data (e.g. genetic sequencing data) to social determinants of health (SDOH) to patient-generated health data. Also new is that, unlike most diagnostic tests where a result interpretation is stable over time, the interpretation of genomic test result data is constantly evolving as new knowledge about the clinical significance of variants is expanding.
Elimu’s vision for Precision Medicine is to leverage this full spectrum of patient-specific data for advanced clinical decision making and ensure that as knowledge about genomic test result data evolves, patients and clinicians are updated when such evolution carries clinical management significance. In prior blogs we’ve talked about FHIR-based strategies to leverage this growing data set for precision medicine. Here, we’ll focus on genetic sequencing data, and emerging FHIR-based strategies for exposing a person’s entire genome for clinical decision support (CDS). In a future blog, we will talk about automated knowledge maintenance strategies, such as the strategy we used under Sync for Genes. Soon, genomics-EHR integration will mean far more than pushing a static PDF report into the EHR, and we can expect to see real-time CDS with rules dynamically updated to reflect current knowledge.
Challenges with Precision Medicine
Challenges with leveraging a person’s entire genome for CDS include:
- Data volume: Genomic data are voluminous and complex. Next-generation sequencing (NGS) can identify thousands to millions of variants.
- Limited interoperability: Raw sequencing data formats and EHR data standards differ, with only a limited number of clinical informaticists having a detailed understanding of bioinformatics data specifications. Significant bioinformatics knowledge and computational resources can be required to make sense of genomic data.
- Rapid evolution of knowledge: The implications of genomic data are rapidly changing as our knowledge matures, resulting in clinically relevant disease and treatment associations quickly becoming outdated.
Today’s EHRs are generally not equipped to deal with a person’s full genetic data set. Further, the rules engines in most EHRs that underlie their clinical decision support functionality are designed to reason over evolving patient data but not evolving clinical knowledge. To manage such a large volume of dynamic and complex data, and to expose that data for CDS, new models of genomics-EHR integration are needed.
Our approach to genomics-EHR integration
Elimu is committed to driving computational advances in precision medicine. We are actively engaged in FHIR Genomics, GenomeX, and GA4GH standards activities and have developed open source solutions for VCF to FHIR translation, for implementing FHIR Genomics Operations, and for demonstrating how FHIR Genomics Operations can dynamically annotate variants with GA4GH-encoded knowledge.
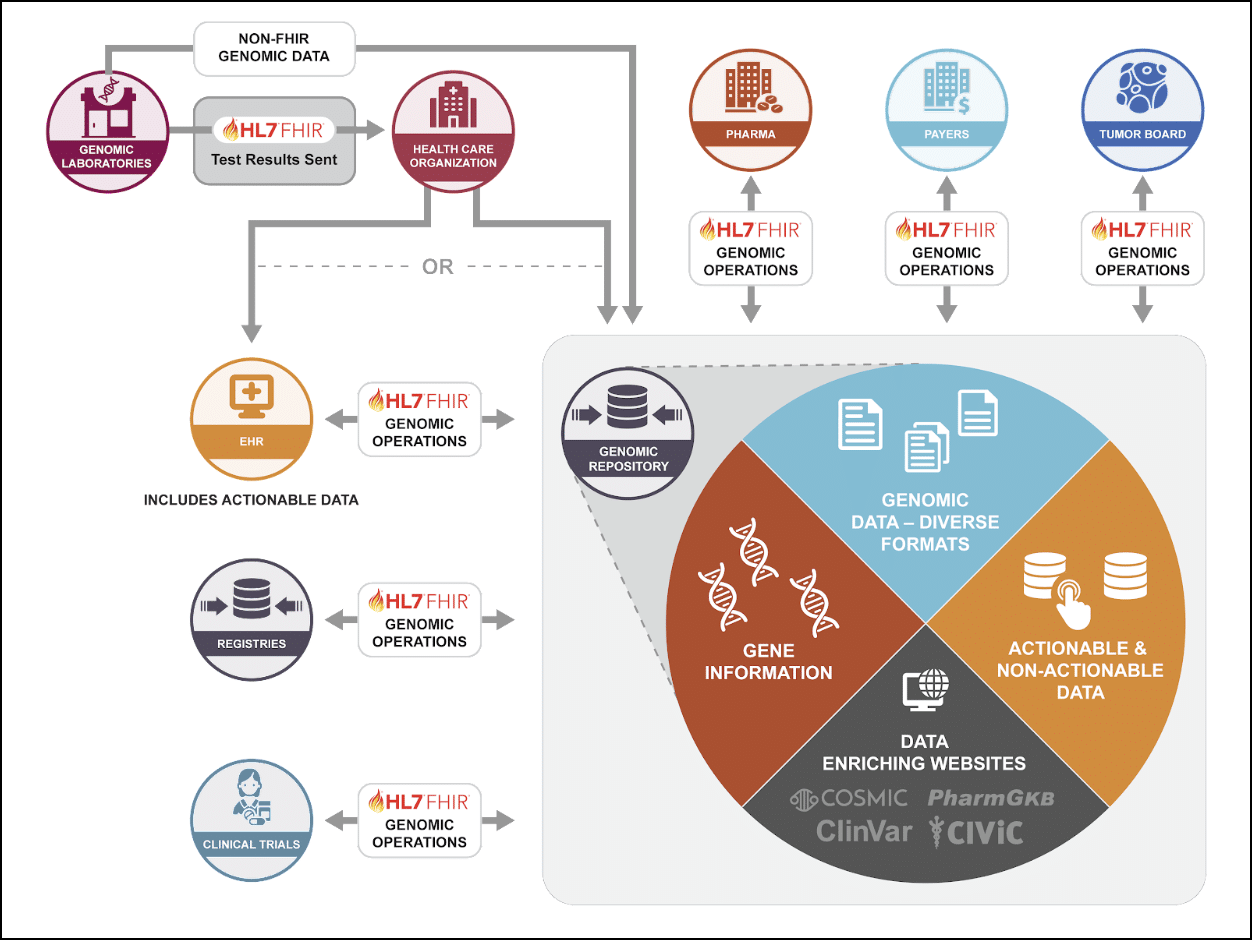
This picture, courtesy of HL7 FHIR Accelerator CodeXTM, illustrates how a genomic repository (aka ‘Genome Archiving and Communication System’ or ‘GACS’) can store raw genomic data and serve up that data via standardized FHIR Genomics Operations, as a complement to existing data interfaces that communicate structured test results directly with the EHR. Note also that a GACS server can store ‘knowledge’, such as knowledge about the implications of genetic findings, including diagnostic and therapeutic implications and predicted molecular consequences. FHIR Genomics Operations can dynamically intersect a person’s genetic findings with this knowledge to return not only genetic findings, but also, the current implications of those findings.
Our team is developing a precision medicine platform aligned with this model that enables FHIR-based strategies for exposing a person’s entire genome for CDS. Primary components include (1) GACS: An evolving prototype based on exploratory storage of various data structures including raw VCF data, chip-based data, vector-encoded genomic data, and other formats; along with knowledge regarding variant implications; (2) SapphireⓇ Omnibus Platform: Our central decision support and workflow engine, with full support for FHIR APIs and FHIR Genomics Operations; (3) SapphireⓇ App Assembler: A SMART-on-FHIR app assembler that is often coupled with Omnibus to achieve advanced decision support and EHR integration capabilities.
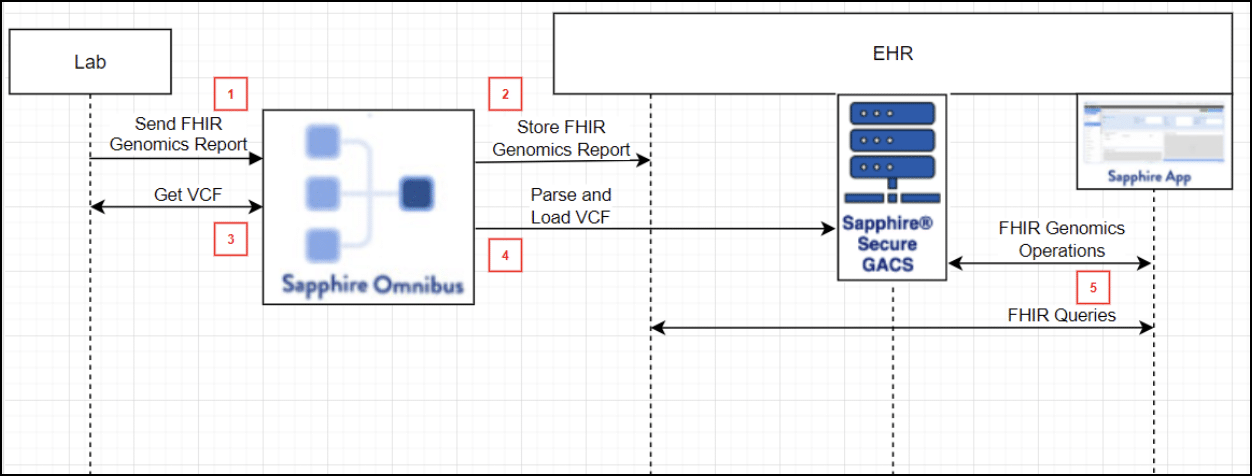
Automating the process of moving non-FHIR Genomic Data (e.g. VCF files) from the lab into GACS so that they can be served up by FHIR Genomics Operations, thereby enabling CDS against a person’s whole genome, is an active area of study. We illustrate our approach in the following figure.
In this approach:
- A lab sends a FHIR Genomics Report to the EHR. Within that report is an explicit reference to raw data files, such as VCF.
- A Sapphire Omnibus service receives the FHIR Genomics Report, stores it in the EHR, and extracts references to raw data files.
- The Sapphire Omnibus service retrieves raw data files from the lab.
- The Sapphire Omnibus service parses raw data files and loads them into GACS.
- Apps can now access data stored in a FHIR server using standard FHIR queries, and can access a patient’s complete genomic data, stored in GACS, using FHIR Genomics Operations.
We are testing this approach with a number of apps (molecular tumor board, rare disease / variant discovery, face sheet, pharmacogenomics, and others).
We are curious to learn more about your genomics-EHR integration use cases, and see if our precision medicine solutions might be the right fit for you. In addition, our team offers advisory services and custom software solutions to help drive genomics-EHR integration and genomics CDS. On top of that, our nerds would welcome a conversation with your nerds, if only to share ideas about where we are going with genomics-EHR integration.